11:50 (13.02.2024)
1958
Using AI, MIT researchers identify a new class of antibiotic candidates
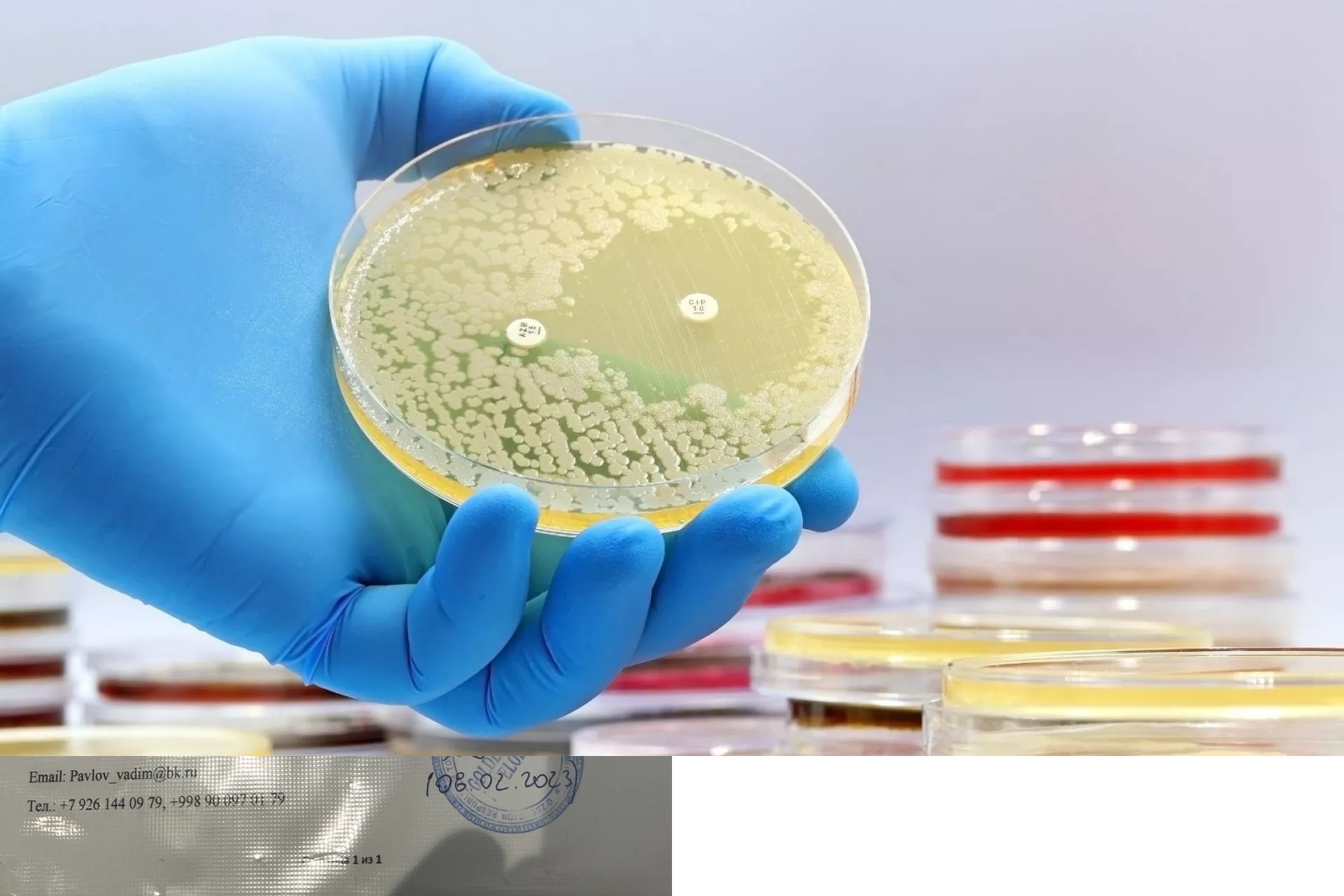
Using a type of artificial intelligence known as deep learning, MIT researchers have discovered a class of compounds that can kill a drug-resistant bacterium that causes more than 10,000 deaths in the United States every year.
In a study appearing today in Nature, the researchers showed that these compounds could kill methicillin-resistant Staphylococcus aureus (MRSA) grown in a lab dish and in two mouse models of MRSA infection. The compounds also show very low toxicity against human cells, making them particularly good drug candidates.
A key innovation of the new study is that the researchers were also able to figure out what kinds of information the deep-learning model was using to make its antibiotic potency predictions. This knowledge could help researchers to design additional drugs that might work even better than the ones identified by the model.
“The insight here was that we could see what was being learned by the models to make their predictions that certain molecules would make for good antibiotics. Our work provides a framework that is time-efficient, resource-efficient, and mechanistically insightful, from a chemical-structure standpoint, in ways that we haven’t had to date,” says James Collins, the Termeer Professor of Medical Engineering and Science in MIT’s Institute for Medical Engineering and Science (IMES) and Department of Biological Engineering.
Felix Wong, a postdoc at IMES and the Broad Institute of MIT and Harvard, and Erica Zheng, a former Harvard Medical School graduate student who was advised by Collins, are the lead authors of the study, which is part of the Antibiotics-AI Project at MIT. The mission of this project, led by Collins, is to discover new classes of antibiotics against seven types of deadly bacteria, over seven years.
MRSA, which infects more than 80,000 people in the United States every year, often causes skin infections or pneumonia. Severe cases can lead to sepsis, a potentially fatal bloodstream infection.
Over the past several years, Collins and his colleagues in MIT’s Abdul Latif Jameel Clinic for Machine Learning in Health (Jameel Clinic) have begun using deep learning to try to find new antibiotics. Their work has yielded potential drugs against Acinetobacter baumannii, a bacterium that is often found in hospitals, and many other drug-resistant bacteria.
These compounds were identified using deep learning models that can learn to identify chemical structures that are associated with antimicrobial activity. These models then sift through millions of other compounds, generating predictions of which ones may have strong antimicrobial activity.
Experiments revealed that the compounds appear to kill bacteria by disrupting their ability to maintain an electrochemical gradient across their cell membranes. This gradient is needed for many critical cell functions, including the ability to produce ATP (molecules that cells use to store energy). An antibiotic candidate that Collins’ lab discovered in 2020, halicin, appears to work by a similar mechanism but is specific to Gram-negative bacteria (bacteria with thin cell walls). MRSA is a Gram-positive bacterium, with thicker cell walls.
“We have pretty strong evidence that this new structural class is active against Gram-positive pathogens by selectively dissipating the proton motive force in bacteria,” Wong says. “The molecules are attacking bacterial cell membranes selectively, in a way that does not incur substantial damage in human cell membranes. Our substantially augmented deep learning approach allowed us to predict this new structural class of antibiotics and enabled the finding that it is not toxic against human cells.”
The researchers have shared their findings with Phare Bio, a nonprofit started by Collins and others as part of the Antibiotics-AI Project. The nonprofit now plans to do more detailed analysis of the chemical properties and potential clinical use of these compounds. Meanwhile, Collins’ lab is working on designing additional drug candidates based on the findings of the new study, as well as using the models to seek compounds that can kill other types of bacteria.